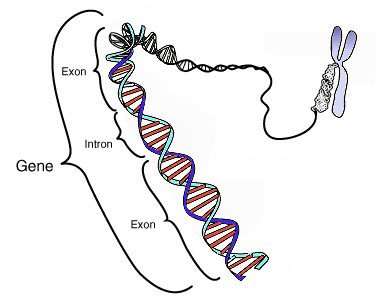
https://upload.wikimedia.org/wikipedia/commons/thumb/3/38/Protein_primary_structure.svg/2000px-Protein_primary_structure.svg.png
Deletion and insertion seemed to be the most changing forms of mutation. Substitution effected the protein code the least. Where the mutation occurs does matter for all the forms of mutation. The earlier the code, the most damage deletion and insertion do. Substitution just need to change an essential nucleotide to make any sort of difference.

https://upload.wikimedia.org/wikipedia/commons/5/50/Mutation_par_substitution.png
I chose the insertion mutation because it seemed to have changed the amino acids the most. Insertion did change the amino acids almost as much as deletion did, and it definitely changed more than substitution. Since I inserted a new nucleotide in the very beginning, it had the biggest effect on the sequence, showing how important where the mutation occurs is.

https://upload.wikimedia.org/wikipedia/commons/thumb/6/6d/RNA-codons-aminoacids.svg/2000px-RNA-codons-aminoacids.svg.png
Mutations could greatly affect my life. They could constrict me from doing many things and functioning properly, or they could be minor and not have to big of an effect on my life. For example, huntington disease is an inherited mutation in which nerve cells in the brain break down over time. Huntington's disease is an autosomal dominant gene. There is a 50% of children inheriting the gene.

http://hdsa.org/wp-content/themes/hdsa/images/img_HD2.png